1. Regulation of Ferritin, Antioxidant Genes, and Oxidative Stress
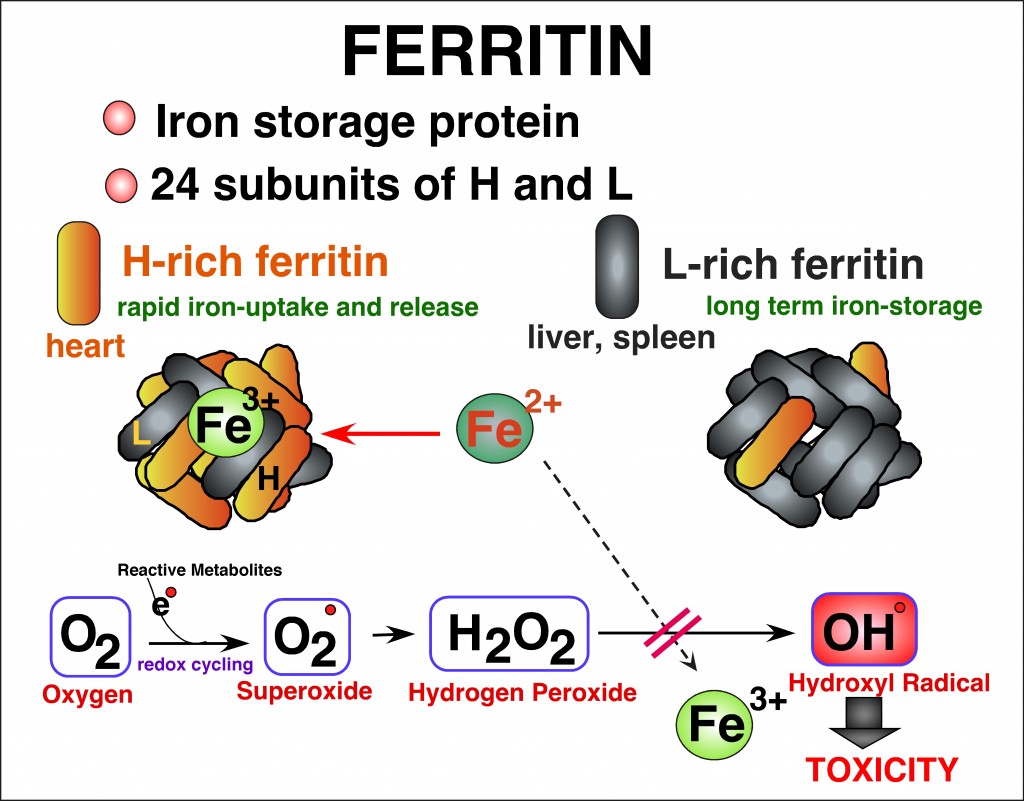
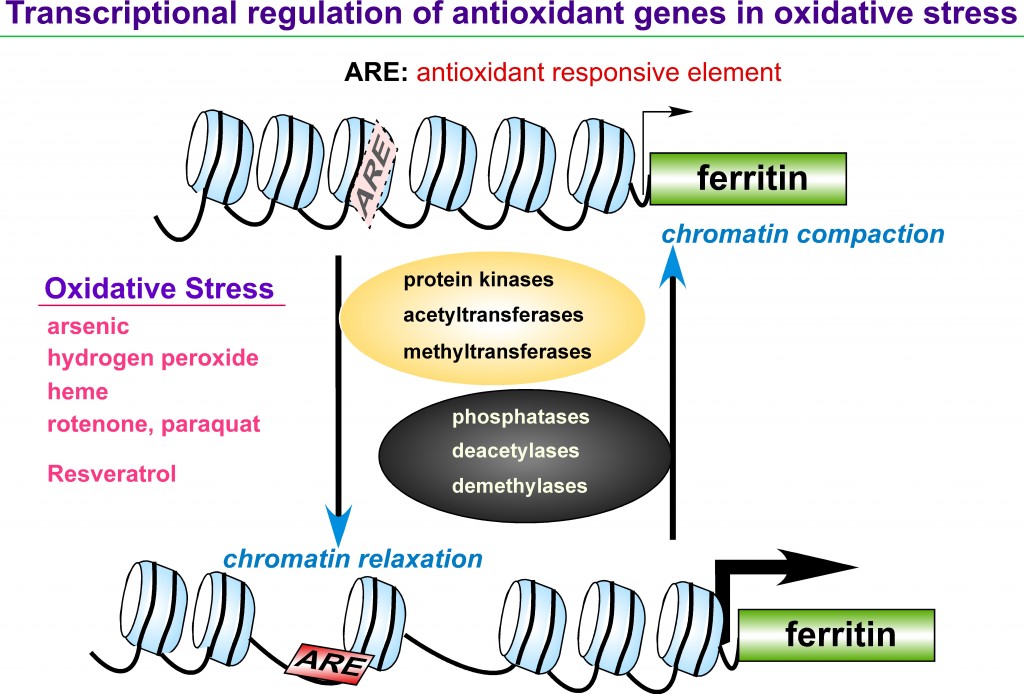
Iron is an essential element in a variety of cellular functions; however, excess iron is detrimental because it catalyzes formation of reactive oxygen species (ROS). Disorders of iron homeostasis involving iron deficiency or overload cause various human health problems such as neurodegenerative disease, cancer and aging. Fine-tuning of intracellular iron levels is therefore essential for maintaining normal cellular function and physiological metabolic balance. Ferritin is the major iron-storage protein in eukaryotic cells and it plays a crucial role in regulation of iron metabolism by detoxifying and storing intracellular excess iron in a non-toxic but bioavailable form. Ferritin synthesis is regulated at both transcriptional and translational levels. Translational regulatory mechanism of ferritin by iron has been extensively studied and well characterized. In contrast, iron-independent transcriptional regulation of the ferritin gene, particularly through chromatin remodeling under oxidative stress conditions, remain incompletely understood. Transcription of ferritin and a battery of antioxidant genes are regulated by a conserved enhancer, termed the ARE (antioxidant responsive element). We hypothesize that chromatin remodeling and associated factors we recently identified on the human ferritin ARE can serve as crucial proteins that regulate ferritin transcription and iron homeostasis. We focus on characterization of these new ARE-interacting proteins and their roles in chromatin modifications adjacent to ARE-regulated ferritin and antioxidant genes. It will define new regulatory proteins responsible for cellular antioxidant response and iron homeostasis under oxidative stress conditions that are associated with various iron- and ROS-involving human diseases.
2. The Role of CREB/ATF1 in Cellular Genotoxic Response to Xenobiotics
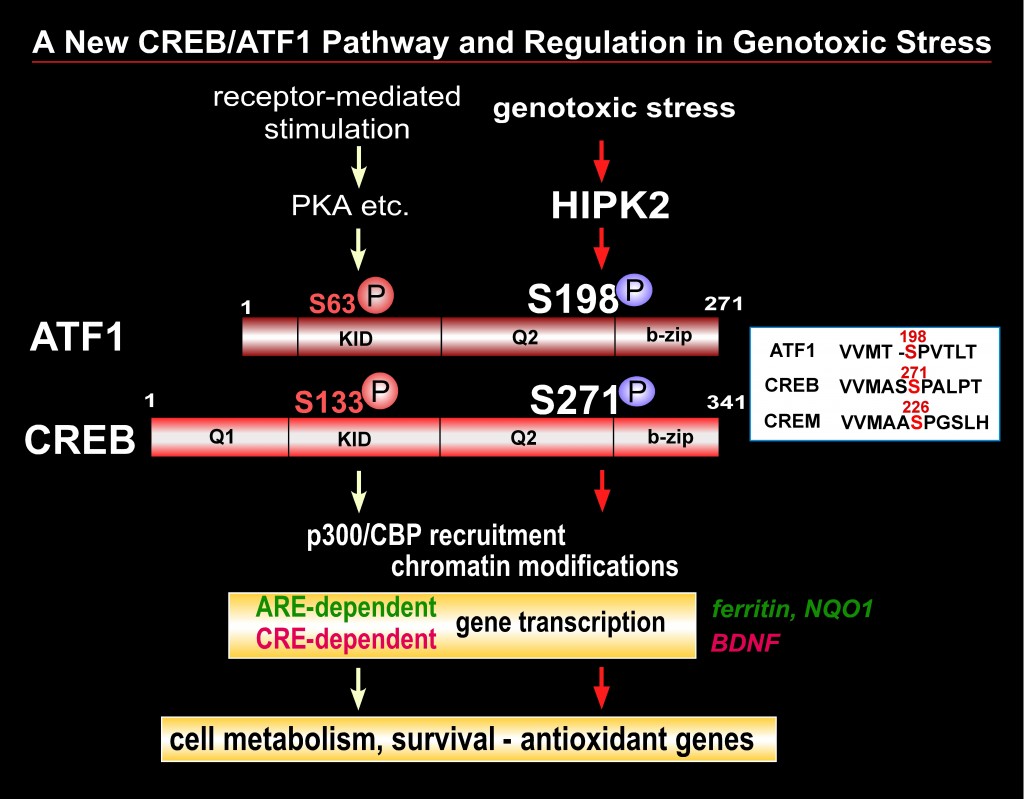
The CREB (cAMP-response element binding) transcription factor is a stimulus-induced phospho-protein that is involved in numerous cellular signaling pathways. Dysfunction and deregulation of CREB and CREB-interacting proteins cause human diseases such as cancer and neurodegeneration. CREB appears to play a key role in cell defense and survival in various tissues; however, the mechanisms through which CREB is involved in cell survival and the reason why deregulation of CREB function causes these human diseases remain incompletely understood. CREB phosphorylation at Ser-133 is the major posttranslational modification that enhances CREB activity in response to receptor-coupled stimuli. We recently found that HIPK2 (homeodomain interacting protein kinase 2), a genotoxic stress responsive kinase, activates CREB via phosphorylation of a new serine site (Ser-271) but not Ser-133. We currently investigate whether HIPK2 is a new regulator of the CREB transcription factor that induces a cell survival program in genotoxic and oxidative stress conditions. We will focus on characterization of molecular mechanism through which CREB phosphorylation by HIPK2 regulates its transcription function as well as downstream events including expression of target genes and cellular susceptibility to genotoxic stress. This project will enhance our understanding in various physiological and disease conditions closely associated with the status of CREB activity.
3. Regulation and Function of p66Shc
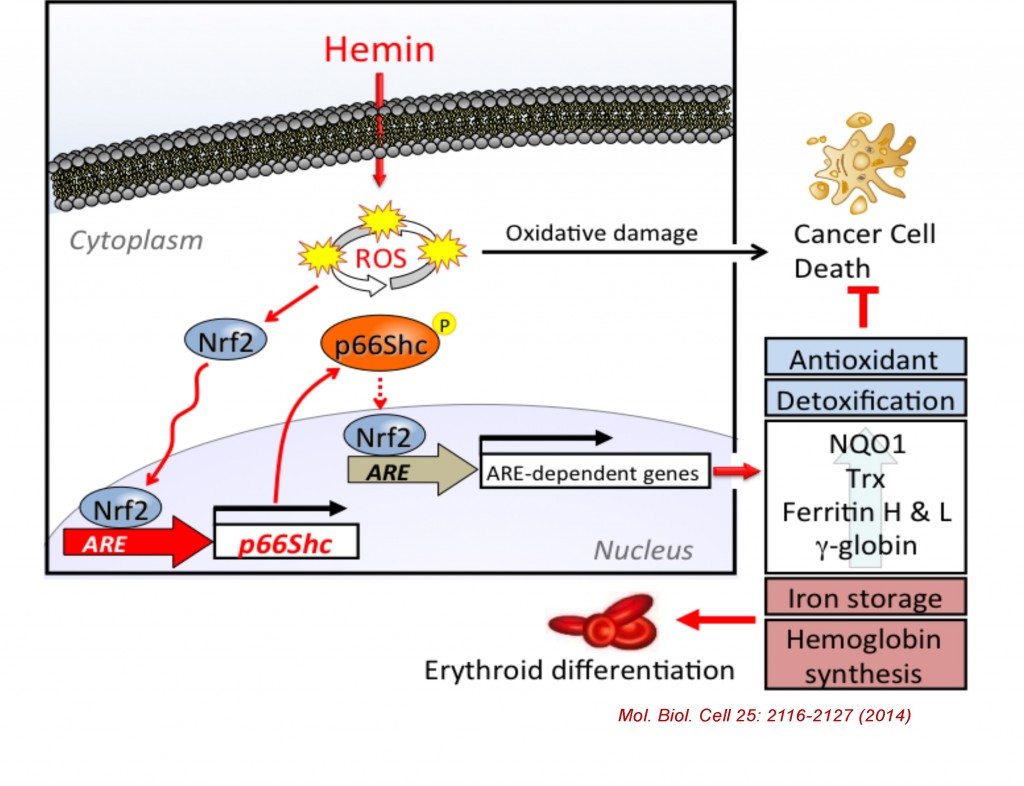
The mammalian Shc family is composed of p46, p52, and p66 isoforms. p46 and p52 Shc isoforms are synthesized from alternative translational initiation site and both serving as adaptor proteins in growth factor signaling pathways. In contrast, p66Shc is regulated by an alternative promoter, allowing p66Shc to gain a unique N-terminal domain and distinct functions from p46Shc and p52Shc. p66Shc knockout mice were shown to extend their life spans by 30% compared to control, suggesting p66Shc serving as a negative lifespan regulatory protein. p66Shc is understood to produce reactive oxygen species in mitochondria therefore plays a prooxidant role. However, we recently found that p66Shc, but not p52Shc or p46Shc, is transcriptionally activated by NFE2-related factor 2 (Nrf2) in K562 and several human cell types following treatment with Nrf2 activators such as hemin (iron-containing porphyrin), sulforaphane, and arsenite. We have recently identified a functional antioxidant response element (ARE) in the 5’ regulatory region of the human p66Shc gene and its interaction with Nrf2. Since Nrf2-regulated genes are generally involved in cellular antioxidant defense system, our results suggest that p66Shc is an antioxidant gene rather than a prooxidant gene. Indeed, p66Shc knockdown as well as Nrf2 knockdown significantly enhanced ROS production along with decreased expression of other ARE-regulated antioxidant genes in K562 cells. We elucidate the new regulatory mechanism of p66Shc expression in various types of cells and challenge the current paradigm of p66Shc function in oxidative stress and aging.
Key Words
ferritin, antioxidant responsive element (ARE), CREB/ATF1, Nrf2, b-zip transcription factors, HIPK2, arsenic, p66Shc, reactive oxygen species (ROS), frataxin, microRNA, histones, lamins